TotalDepth Command Line Tools¶
This describes the command line tools that are available for processing any TotalDepth file.
The tools are located in TotalDepth/
| Tool Name | Description |
|---|---|
tdarchive |
Scans a directory of files and provides an analysis by file type. Link |
tdcopybinfiles |
Selects files of a particular type and copies them to a directory. This can also expand compressed archives. Link |
tdremovedupefiles |
Removes duplicate files based on their checksum. Link |
tddetif |
Removes TIF markers from a file. Link |
tdplotlogs |
Plots logs from LIS and LAS data. Link |
Analysing a Directory of Files with tdarchive¶
Scans a directory of files and provides an analysis by file type.
Usage¶
Usage:
usage: tdarchive [-h] [--file-type FILE_TYPE] [-b BYTES] [-r]
[--expand-and-delete] [--histogram] [-n] [-o] [-l LOG_LEVEL]
[-v]
path [path_out]
Arguments¶
- The path to the input directory
- [Optional] Path to the output directory to write the files to. The results are undefined if path_out conflicts with path_in.
Options¶
| Option | Description |
|---|---|
-h, --help |
Show this help message and exit. |
--file-type |
Types of files to analyse, this option is is additive so can be used multiple times. Supported (and default) file types are: ASCII, LAS1.2, LAS2.0, LAS3.0, LIS, LISt, LIStr, PDF, PS, RP66V1, RP66V1t, RP66V1tr, RP66V2, SEGY, TIFF, XML, ZIP |
-b, --bytes= |
The number of initial bytes of the file to show [default: 0]. |
-r, --recursive |
Process input recursively. [default: False] |
--expand-and-delete |
Expand and delete archive files, implies --recurse. [default: False] |
--histogram |
Plot a histogram of file sizes. [default: False] |
-n, --nervous |
Nervous mode, does not do anything but report. |
-o, --over-write |
Overwrite existing files, otherwise warns. |
-l, --log-level= |
Log Level as an integer or symbol. (0<->NOTSET, 10<->DEBUG, 20<->INFO, 30<->WARNING, 40<->ERROR, 50<->CRITICAL) [default: 30] |
-v, --verbose |
Increase verbosity, additive [default: 0] |
Examples¶
Here is an example of scanning the example_data directory (the output is filleted for clarity):
$ tdarchive example_data/ -r --histogram
CMD: tdarchive example_data/ -r --histogram
Analysing archive.
Common prefix:
108,707 .SVG XML example_data/LIS/Plot/DILLSON-1_WELL_LOGS_FILE-013.LIS_0001_HDT.svg
667 .HTML XML example_data/LIS/Plot/index.html
163,169 .SVG XML example_data/LIS/Plot/DILLSON-1_WELL_LOGS_FILE-037.LIS_0001_1.svg
690 .CSS ASCII example_data/LIS/Plot/index.css
...
184,084 .LIS LIS example_data/LIS/data/DILLSON-1_WELL_LOGS_FILE-037.LIS
96,376 .LIS LIS example_data/LIS/data/DILLSON-1_WELL_LOGS_FILE-013.LIS
98,508 .LIS LIS example_data/LIS/data/DILLSON-1_WELL_LOGS_FILE-049.LIS
10,306 .PY ASCII example_data/RP66V1/demo_read.py
462,795 .HTML XML example_data/RP66V1/HTML/206_05a-_3_DWL_DWL_WIRE_258276498.DLIS.HTML.html
3,098 .HTML XML example_data/RP66V1/HTML/index.html
...
1,018,327 .PKL example_data/RP66V1/pickle/206_05a-_3_DWL_DWL_WIRE_258276498.pkl
276,045 .PKL example_data/RP66V1/pickle/206_05a-_3_DWL_DWL_WIRE_258276498.DLIS.pkl
520 .PKL example_data/RP66V1/pickle/MINIMAL_FILE.dlis.pkl
53,137 .PKL example_data/RP66V1/pickle/BASIC_FILE.dlis.pkl
949 .PKL example_data/RP66V1/pickle/BASIC_FILE_WITH_TWO_VISIBLE_RECORDS_NO_IFLRS.dlis.pkl
7,731 .LAS LAS2.0 example_data/RP66V1/LAS/BASIC_FILE_WITH_TWO_VISIBLE_RECORDS_NO_IFLRS_0_.las
1,621,374 .LAS LAS2.0 example_data/RP66V1/LAS/206_05a-_3_DWL_DWL_WIRE_258276498_0_800T.las
...
44,916 .DLIS RP66V1 example_data/RP66V1/data/BASIC_FILE.dlis
540,372 .DLIS RP66V1 example_data/RP66V1/data/206_05a-_3_DWL_DWL_WIRE_258276498.DLIS
716 .DLIS RP66V1 example_data/RP66V1/data/MINIMAL_FILE.dlis
8,826 .DLIS RP66V1 example_data/RP66V1/data/BASIC_FILE_WITH_TWO_VISIBLE_RECORDS_NO_IFLRS.dlis
Total number of files 75, total bytes 12,981,766
File extensions:
.CSS : 5
.DLIS : 4
.HTML : 13
.LAS : 6
.LIS : 3
.PKL : 5
.PY : 1
.PYC : 1
.SVG : 36
.XML : 1
Binary file types:
Binary type: ""
Extensions: .PKL, .PYC
Count: 6 [8.000%]
Bytes: 1,354,629 [10.435%] from 520 to 1,018,327
>=2**9 [ 2] | ++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
>=2**10 [ 0] |
>=2**11 [ 0] |
>=2**12 [ 1] | ++++++++++++++++++++++++++++++++++++++++
>=2**13 [ 0] |
>=2**14 [ 0] |
>=2**15 [ 1] | ++++++++++++++++++++++++++++++++++++++++
>=2**16 [ 0] |
>=2**17 [ 0] |
>=2**18 [ 1] | ++++++++++++++++++++++++++++++++++++++++
>=2**19 [ 1] | ++++++++++++++++++++++++++++++++++++++++
Binary type: "ASCII"
Extensions: .CSS, .PY
Count: 6 [8.000%]
Bytes: 15,232 [0.117%] from 690 to 10,306
>=2**9 [ 4] | ++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
>=2**10 [ 0] |
>=2**11 [ 1] | ++++++++++++++++++++
>=2**12 [ 0] |
>=2**13 [ 1] | ++++++++++++++++++++
Binary type: "LAS2.0"
Extensions: .LAS
Count: 6 [8.000%]
Bytes: 1,785,653 [13.755%] from 1,279 to 1,621,374
>=2**10 [ 1] | ++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
>=2**11 [ 1] | ++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
>=2**12 [ 1] | ++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
>=2**13 [ 0] |
>=2**14 [ 0] |
>=2**15 [ 1] | ++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
>=2**16 [ 1] | ++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
>=2**17 [ 0] |
>=2**18 [ 0] |
>=2**19 [ 0] |
>=2**20 [ 1] | ++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
Binary type: "LIS"
Extensions: .LIS
Count: 3 [4.000%]
Bytes: 378,968 [2.919%] from 96,376 to 184,084
>=2**16 [ 2] | ++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
>=2**17 [ 1] | ++++++++++++++++++++++++++++++++++++++++
Binary type: "RP66V1"
Extensions: .DLIS
Count: 4 [5.333%]
Bytes: 594,830 [4.582%] from 716 to 540,372
>=2**9 [ 1] | ++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
>=2**10 [ 0] |
>=2**11 [ 0] |
>=2**12 [ 0] |
>=2**13 [ 1] | ++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
>=2**14 [ 0] |
>=2**15 [ 1] | ++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
>=2**16 [ 0] |
>=2**17 [ 0] |
>=2**18 [ 0] |
>=2**19 [ 1] | ++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
Binary type: "XML"
Extensions: .HTML, .SVG, .XML
Count: 50 [66.667%]
Bytes: 8,852,454 [68.191%] from 667 to 961,863
>=2**9 [ 1] | ++++
>=2**10 [ 1] | ++++
>=2**11 [ 2] | +++++++++
>=2**12 [ 2] | +++++++++
>=2**13 [ 0] |
>=2**14 [ 0] |
>=2**15 [ 3] | +++++++++++++
>=2**16 [ 16] | +++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
>=2**17 [ 18] | ++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
>=2**18 [ 5] | ++++++++++++++++++++++
>=2**19 [ 2] | +++++++++
Execution time: 0.067 (s)
Files: 75 rate 1,113.2 (files/s)
Bytes: 1,350 rate 20,038.5 (bytes/s)
Copying a Directory of Files with tdcopybinfiles¶
Scans a directory of files and can copy particular file type to another directory.
Usage¶
Usage:
usage: TotalDepth.RP66V1.util.CopyBinFiles.main
[-h] [--file-types FILE_TYPES] [-m] [-n] [-l LOG_LEVEL]
path_in path_out
Arguments¶
- The path to the input directory
- Path to the output directory to write the files to. The results are undefined if path_out conflicts with path_in.
Options¶
| Option | Description |
|---|---|
-h, --help |
Show this help message and exit. |
--file-type |
Types of files to copy, this option is is additive so can be used multiple times. Supported (and default) file types are: ASCII, LAS1.2, LAS2.0, LAS3.0, LIS, LISt, LIStr, PDF, PS, RP66V1, RP66V1t, RP66V1tr, RP66V2, SEGY, TIFF, XML, ZIP Use ‘?’ or ‘??’ to see what file types are available |
-m, --move |
Move rather than copy, Irrelevant for files in ZIP archives which are always copied. [default: False] |
-n, --nervous |
Nervous mode, does not do anything but report. |
-l, --log-level= |
Log Level as an integer or symbol. (0<->NOTSET, 10<->DEBUG, 20<->INFO, 30<->WARNING, 40<->ERROR, 50<->CRITICAL) [default: 30] |
Examples¶
To see what file types are supported use ‘?’ and two dummy paths:
$ tdcopybinfiles --file-types=? '' ''
Cmd: tdcopybinfiles --file-types=?
Binary file types supported: ASCII, LAS1.2, LAS2.0, LAS3.0, LIS, LISt, LIStr, PDF, PS, RP66V1, RP66V1t, RP66V1tr, RP66V2, SEGY, TIFF, XML, ZIP
Execution time = 0.000 (S) 0 kb/s
To get the file type description as well use ‘??’ and two dummy paths:
$ tdcopybinfiles --file-types=?? '' ''
Cmd: tdcopybinfiles --file-types=??
Binary file types supported:
ASCII - American Standard Code for Information Interchange
LAS1.2 - Canadian Well Logging Society Log ASCII Standard version 1.2
LAS2.0 - Canadian Well Logging Society Log ASCII Standard version 2.0
LAS3.0 - Canadian Well Logging Society Log ASCII Standard version 3.0
LIS - Schlumberger LIS-79 well logging format
LISt - Schlumberger LIS-79 well logging format with TIF markers
LIStr - Schlumberger LIS-79 well logging format with reversed TIF markers
PDF - Portable Document Format
PS - Postscript
RP66V1 - American Petroleum Institute Recommended Practice 66 version 1
RP66V1t - American Petroleum Institute Recommended Practice 66 version 1 with TIF markers
RP66V1tr - American Petroleum Institute Recommended Practice 66 version 1 with reversed TIF markers
RP66V2 - American Petroleum Institute Recommended Practice 66 version 2
SEGY - Society of Exploration Geophysicists seismic format Y
TIFF - Tagged Image File Format
XML - eXtensible Markup Language
ZIP - ZIP Compressed Archive
Execution time = 0.000 (S) 0 kb/s
Removing Duplicate Files with tdremovedupefiles¶
Scans a directory of files and identifies duplicate files by their checksum.
It is strongly recommended to use -n (--nervous) first and look at the results before running this without -n which is potentially destructive.
Usage¶
Usage:
usage: TotalDepth.RP66V1.util.RemoveDupeFiles.main [-h] [--version] [-k] [-v]
[-r] [-l LOG_LEVEL] [-n]
path_in
Arguments¶
- The path to the directory.
Options¶
| Option | Description |
|---|---|
-h, --help |
Show this help message and exit. |
-r, --recursive |
Process input recursively. [default: False] |
-n, --nervous |
Nervous mode, does not do anything but report [default: False]. |
-l, --log-level= |
Log Level as an integer or symbol. (0<->NOTSET, 10<->DEBUG, 20<->INFO, 30<->WARNING, 40<->ERROR, 50<->CRITICAL) [default: 20] |
-v, --verbose |
Increase verbosity, additive [default: 0] |
Removing TIF Markers From Files with tddetif¶
Scans a directory of files and removes TIF markers. TIF markers are 12 bytes of data inserted in various places. They are not part of any standard and, except for one case, provide no value. This tool rewrites the input file without TIF markers if they are found.
Arguments¶
- The path to the input directory
- Path to the output directory to write the files to. The results are undefined if path_out conflicts with path_in.
Options¶
| Option | Description |
|---|---|
-h, --help |
Show this help message and exit. |
-r, --recurse |
Process input recursively. [default: False] |
-n, --nervous |
Nervous mode, does not do anything but report [default: False]. |
-l, --log-level= |
Log Level as an integer or symbol. (0<->NOTSET, 10<->DEBUG, 20<->INFO, 30<->WARNING, 40<->ERROR, 50<->CRITICAL) [default: 30] |
-v, --verbose |
Increase verbosity, additive [default: 0] |
-o, --over-write |
Overwrite existing files if found, otherwise warns of existing target file. |
Plotting Well Logs with tdplotlogs¶
Produces SVG plots from LIS and LAS files.
Usage¶
Usage:
usage: tdplotlogs [-h] [--version] [-j JOBS] [-k] [-l LOGLEVEL] [-g] [-r]
[-A] [-x LGFORMAT] [-X LGFORMAT_MIN] [-s SCALE]
in out
Arguments¶
These are required arguments unless -h or --version options are specified (in which case no processing is done):
- The path to the input LAS or LIS file or directory thereof.
- The path to the output SVG file or directory, any directories will be created as necessary.
Options¶
| Option | Description |
|---|---|
--version |
Show program’s version number and exit |
-h, --help |
Show this help message and exit. |
-j JOBS, --jobs=JOBS |
Max processes when multiprocessing. Zero uses number of native CPUs [8]. -1 disables multiprocessing. [default: -1] |
-k, --keep-going |
Keep going as far as sensible. [default: False] |
-l LOGLEVEL, --loglevel=LOGLEVEL |
Log Level (debug=10, info=20, warning=30, error=40, critical=50) [default: 20] |
-g, --glob |
File pattern match. [default none] |
-r, --recursive |
Process input recursively. [default: False] |
-A, --API |
Put an API header on each plot. [default: False] |
-x LGFORMAT, --xml LGFORMAT |
Use XML LgFormat UniqueId to use for plotting (additive). Use -x? to see what LgFormats (UniqueID+Description) are available. Use -x?? to see what curves each format can plot. See also -X. This is additive so can used multiple times to get multiple plots from the same data. |
-X LGFORMAT_MIN,
--XML LGFORMAT_MIN |
Use all available LgFormat XML plots that use LGFORMAT_MIN or more outputs. If -x option present limited by those LgFormats [default: 0] |
-s SCALE, --scale SCALE |
Scale of X axis to use (an integer). This overrides the scale(s) specified in the LgFormat file or FILM table. [default: 0]. |
Examples¶
LgFormat XML¶
Using -x? to see what formats are available:
$ python3 tdplotlogs -x? spam eggs
The output is something like:
Cmd: tdplotlogs -x? spam eggs
XML LgFormats available: [29]
UniqueId Description
----------------------------------- --------------------------------
ADN_Image_Format : ADN Image Log
Azimuthal_Density_3Track.xml : Azimuthal Density 3Track
Azimuthal_Resistivity_3Track.xml : Azimuthal Resistivity 3Track
Blank_3Track_Depth : Blank 3Track
Blank_3Track_Time.xml : Blank 3Track Time
FMI_IMAGE_ALIGNED : FMI Image Aligned
FMI_IMAGE_PROCESSED : FMI Image Processed
Formation_Test : Formation Test Time
HDT : High Definition Dipmeter
Micro_Resistivity_3Track.xml : Micro Resistivity 3 Track Format
Natural_GR_Spectrometry_3Track.xml : Natural GR Spectrometry 3Track
OBMI_IMAGE_EQUAL : OBMI Image Equalized
Porosity_GR_3Track : Standard Porosity Curves
Pulsed_Neutron_3Track.xml : Pulsed Neutron 3Track
Pulsed_Neutron_Time.xml : Pulsed Neutron Time
RAB_Image_Format_Deep : Resistivity At the Bit Image
RAB_Image_Format_Medium : Resistivity At the Bit Image
RAB_Image_Format_Shallow : Resistivity At the Bit Image
RAB_Std_Format : Resistivity At the Bit
Resistivity_3Track_Correlation.xml : Resistivity Linear Correlation Format
Resistivity_3Track_Logrithmic.xml : Logrithmic Resistivity 3Track
Resistivity_Investigation_Image.xml : AIT Radial Investigation Image
Sonic_3Track.xml : Sonic DT Porosity 3 Track
Sonic_PWF4 : SONIC Packed Waveform 4
Sonic_SPR1_VDL : SONIC Receiver Array Lower Dipole VDL
Sonic_SPR2_VDL : SONIC Receiver Array Upper Dipole VDL
Sonic_SPR3_VDL : SONIC Receiver Array Stonely VDL
Sonic_SPR4_VDL : SONIC Receiver Array P and S VDL
Triple_Combo : Resistivity Density Neutron GR 3Track Format
The first column is the UniqueID to be used in identifying plots for the -x option.
Using -x?? to see what formats and what curves would be plotted by each plot specification:
$ python3 tdplotlogs -x?? a b
The output is something like:
Cmd: tdplotlogs -x?? a b
XML LgFormats available: [29]
UniqueId Description
----------------------------------- --------------------------------
ADN_Image_Format : ADN Image Log
DRHB, GR , GR_RAB, ROBB, ROP5, TNPH
Azimuthal_Density_3Track.xml : Azimuthal Density 3Track
BS , DCAL, DRHB, DRHL, DRHO, DRHR, DRHU, DTAB, HDIA, PEB , PEF , PEL
PER , PEU , RHOB, ROBB, ROBL, ROBR, ROBU, ROP5, RPM , SCN2, SOAB, SOAL
SOAR, SOAU, SONB, SOXB, VDIA
Azimuthal_Resistivity_3Track.xml : Azimuthal Resistivity 3Track
AAI , BS , C1 , C2 , CALI, GR , GRDN_RAB, GRLT_RAB, GRRT_RAB, GRUP_RAB, PCAL, RDBD
RDBL, RDBR, RDBU, RLA0, RLA1, RLA2, RLA3, RLA4, RLA5, RMBD, RMBL, RMBR
RMBU, ROP5, RPM , RSBD, RSBL, RSBR, RSBU, SP , TENS
Blank_3Track_Depth : Blank 3Track
Blank_3Track_Time.xml : Blank 3Track Time
FMI_IMAGE_ALIGNED : FMI Image Aligned
C1 , C2 , GR , HAZIM, P1AZ, SP , TENS
FMI_IMAGE_PROCESSED : FMI Image Processed
C1 , C2 , GR , HAZIM, P1AZ, SP , TENS
Formation_Test : Formation Test Time
B1TR, BFR1, BQP1, BQP1, BQP1, BQP1, BSG1, POHP
HDT : High Definition Dipmeter
C1 , C2 , DEVI, FC0 , FC1 , FC2 , FC3 , FC4 , GR , HAZI, P1AZ, RB
Micro_Resistivity_3Track.xml : Micro Resistivity 3 Track Format
BMIN, BMNO, BS , CALI, GR , HCAL, HMIN, HMNO, MINV, MLL , MNOR, MSFL
PROX, RXO , SP , TENS
Natural_GR_Spectrometry_3Track.xml : Natural GR Spectrometry 3Track
CGR , PCAL, POTA, ROP5, SGR , SIGM, TENS, THOR, URAN
OBMI_IMAGE_EQUAL : OBMI Image Equalized
C1 , C1_OBMT, C2 , C2_OBMT, GR , HAZIM, OBRA3, OBRB3, OBRC3, OBRD3, P1AZ, P1NO_OBMT
TENS
Porosity_GR_3Track : Standard Porosity Curves
APDC, APLC, APSC, BS , C1 , C2 , CALI, CALI_CDN, CMFF, CMRP, DPHB, DPHI
DPHZ, DPOR_CDN, DRHO, ENPH, GR , HCAL, NPHI, NPOR, PCAL, RHOB, RHOZ, ROP5
SNP , SP , SPHI, TENS, TNPB, TNPH, TNPH_CDN, TPHI
Pulsed_Neutron_3Track.xml : Pulsed Neutron 3Track
FBAC, GR , INFD, SIGM, TAU , TCAF, TENS, TPHI, TSCF, TSCN
Pulsed_Neutron_Time.xml : Pulsed Neutron Time
FBAC_SL, GR_SL, INFD_SL, SIGM_SL, TAU_SL, TCAF_SL, TENS_SL, TPHI_SL, TSCF_SL, TSCN_SL
RAB_Image_Format_Deep : Resistivity At the Bit Image
GR_RAB, RES_BD, RES_BM, RES_BS, RES_RING, ROP5
RAB_Image_Format_Medium : Resistivity At the Bit Image
GR_RAB, RES_BD, RES_BM, RES_BS, RES_RING, ROP5
RAB_Image_Format_Shallow : Resistivity At the Bit Image
GR_RAB, RES_BD, RES_BM, RES_BS, RES_RING, ROP5
RAB_Std_Format : Resistivity At the Bit
AAI , BDAV, BDM3, BMAV, BMM2, BSAV, BSM1, BTAB, CALI, DEVI, GR_RAB, HAZI
OBIT, RBIT, RING, ROP5, RPM , RTAB
Resistivity_3Track_Correlation.xml : Resistivity Linear Correlation Format
AHT20, AHT60, AHT90, ATR , BS , CALI, CATR, CILD, CLLD, GR , HCAL, ILD
ILM , LLD , LLS , MSFL, PCAL, PSR , RLA0, ROP5, RT , RXO , SFL , SP
TENS
Resistivity_3Track_Logrithmic.xml : Logrithmic Resistivity 3Track
A22H, A34H, AHF10, AHF20, AHF30, AHF60, AHF90, AHO10, AHO20, AHO30, AHO60, AHO90
AHT10, AHT20, AHT30, AHT60, AHT90, ATR , BS , CALI, GR , HCAL, ILD , ILM
LLD , LLM , MSFL, P16H_RT, P28H_RT, P34H_RT, PCAL, PSR , RLA0, RLA1, RLA2, RLA3
RLA4, RLA5, ROP5, RXO , SFL , SP , TENS
Resistivity_Investigation_Image.xml : AIT Radial Investigation Image
AHT10, AHT20, AHT30, AHT60, AHT90, BS , GR , HCAL, SP
Sonic_3Track.xml : Sonic DT Porosity 3 Track
BS , CALI, DT , DT0S, DT1R, DT2 , DT2R, DT4S, DTBC, DTCO, DTCU, DTL
DTLF, DTLN, DTR2, DTR5, DTRA, DTRS, DTSH, DTSM, DTST, DTTA, GR , HCAL
PCAL, ROP5, SP , SPHI, TENS
Sonic_PWF4 : SONIC Packed Waveform 4
CALI, DT1 , DT2 , DTCO, DTSM, DTST, GR , HCAL, TENS
Sonic_SPR1_VDL : SONIC Receiver Array Lower Dipole VDL
CALI, DT1 , DT1 , DT2 , DTCO, DTSM, DTST, GR , HCAL, TENS
Sonic_SPR2_VDL : SONIC Receiver Array Upper Dipole VDL
CALI, DT1 , DT2 , DT2 , DTCO, DTSM, DTST, GR , HCAL, TENS
Sonic_SPR3_VDL : SONIC Receiver Array Stonely VDL
CALI, DT1 , DT2 , DT3R, DTCO, DTSM, DTST, GR , HCAL, TENS
Sonic_SPR4_VDL : SONIC Receiver Array P and S VDL
CALI, DT1 , DT2 , DTCO, DTRP, DTRS, DTSM, DTST, GR , HCAL, TENS
Triple_Combo : Resistivity Density Neutron GR 3Track Format
AHT10, AHT20, AHT30, AHT60, AHT90, APDC, APLC, APSC, ATR , BS , C1 , C2
CALI, CMFF, CMRP, DPHB, DPHI, DPHZ, DPOR_CDN, DSOZ, ENPH, GR , HCAL, HMIN
HMNO, ILD , ILM , LLD , LLM , MSFL, NPHI, NPOR, PCAL, PEFZ, PSR , RLA0
RLA1, RLA2, RLA3, RLA4, RLA5, ROP5, RSOZ, RXO , RXOZ, SFL , SNP , SP
SPHI, TENS, TNPB, TNPH, TNPH_CDN, TPHI
Plotting Logs¶
Here is an example of plotting LIS and LAS files in directory in/ with the plots in directory out/. The following options have been invoked:
- API headers on the top of each plot:
-A - Multiprocessing on with 4 simultaneous jobs:
-j4 - Recursive search of input directory:
-r - Uses any available plot specifications from LgFormat XML files which result in 4 curves or more being plotted:
-X 4
The command line is:
$ python3 tdplotlogs -A -j4 -r -X 4 in/ out/
First tdplotlogs echos the command:
Cmd: tdplotlogs -A -j4 -r -X 4 in/ out/
When complete tdplotlogs writes out a summary, first the number of files read (output is wrapped here with ‘' for clarity):
plotLogInfo PlotLogInfo <__main__.PlotLogInfo object at 0x101e0da90> \
Files=23 \
Bytes=10648531 \
LogPasses=23 \
Plots=8 \
Curve points=229991
Then as summary of each plot in detail (output is wrapped here with ‘' for clarity):
('in/1003578128.las', \
0, \
'Natural_GR_Spectrometry_3Track.xml', \
IndexTableValue( \
scale=100, \
evFirst='800.5 (FEET)', \
evLast='3019.5 (FEET)', \
evInterval='2219.0 (FEET)', \
curves='CGR_2, POTA, SGR_1, TENS_16, THOR, URAN', \
numPoints=26213, \
outPath='out//1003578128.las_0000_Natural_GR_Spectrometry_3Track.xml.svg' \
)
)
('in/1003578128.las', \
0, \
'Porosity_GR_3Track', \
IndexTableValue( \
scale=100, \
evFirst='800.5 (FEET)', \
evLast='3019.5 (FEET)', \
evInterval='2219.0 (FEET)', \
curves='Cali, DRHO, DensityPorosity, GammaRay, NeutronPorosity, OLDESTNeutronPorosity, OLDNeutronPorosity, RHOB, SP, SonicPorosity, Tension', \
numPoints=46170, \
outPath='out//1003578128.las_0000_Porosity_GR_3Track.svg' \
)
)
... 8<------------- Snip ------------->8
('in/1006346987.las', \
0, 'Sonic_3Track.xml', \
IndexTableValue(
scale=100, \
evFirst='4597.5 (FEET)', \
evLast='5799.5 (FEET)', \
evInterval='1202.0 (FEET)', \
curves='Caliper, DT, DTL_DDBHC, GammaRay, SonicPorosity, TENSION', \
numPoints=14430, \
outPath='out//1006346987.las_0000_Sonic_3Track.xml.svg' \
)
)
The fields in each tuple are:
- Input file name.
- LogPass number in the file. For example “Repeat Section” might be 0 and “Main Log” 1.
- LgFormat used for the plot (several plots my be generated from one LogPass).
- An IndexTableValue object (used to generate the index.html file) that has the following fields:
- Plot scale as an integer.
- First reading and units as an Engineering Value.
- Last reading and units as an Engineering Value.
- Log interval and units as an Engineering Value.
- List of curve names plotted.
- Total number of data points plotted.
- The ouput file.
Finally the total number of curve feet plotted and the time it took:
Interval*curves: EngVal: 121020.000 (FEET)
CPU time = 0.043 (S)
Exec. time = 25.119 (S)
Bye, bye!
In this case (under Unix) the “CPU Time” is the cumulative amount of CPU time used. As we are using multiprocessing it is the CPU time of the parent process which is very small since it just invokes child processes. The Exec. time is the wall clock time between starting and finishing tdplotlogs.
In the output directory will be an index.html file that has a table with the fields that duplicate those on the command line output. It looks like this:
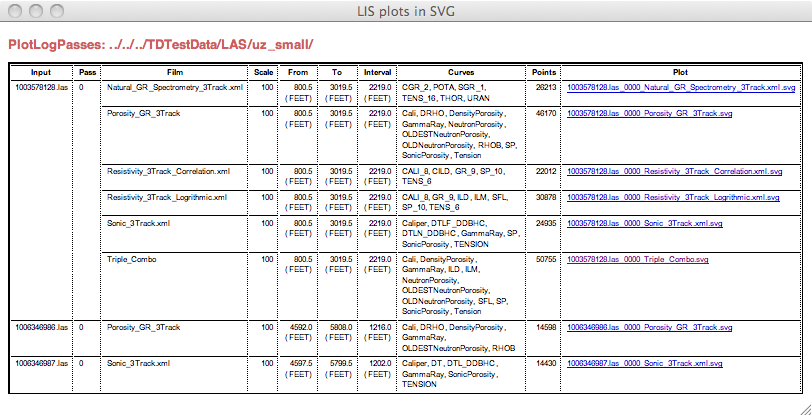
The links in the last column are to the SVG plots. Her is a screen shot of one:
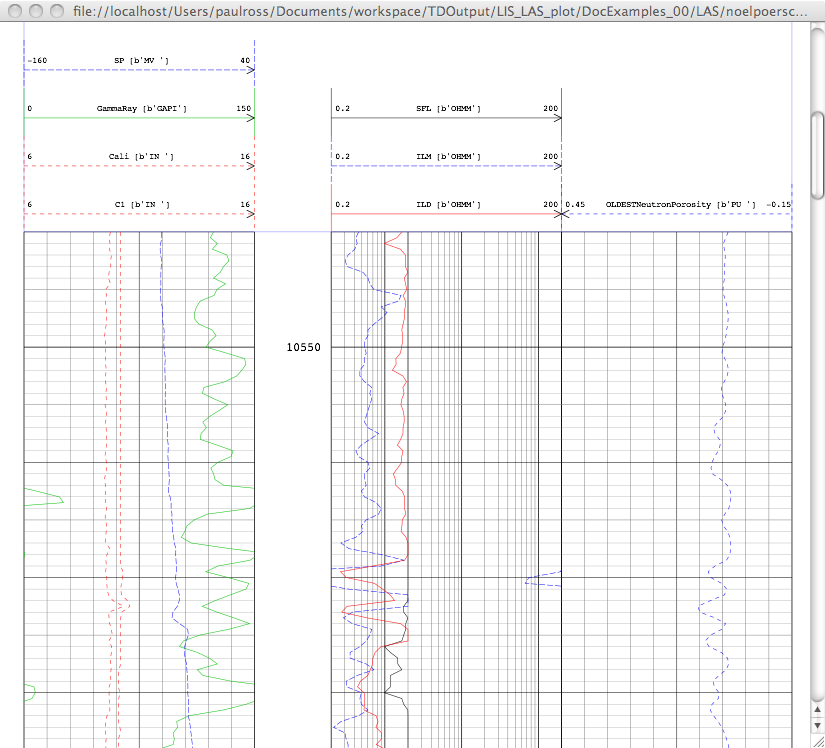
Sample Plots¶
Here is an actual plot from a LAS file and there are many more examples here: Wireline Plots.